The Distributed Networks
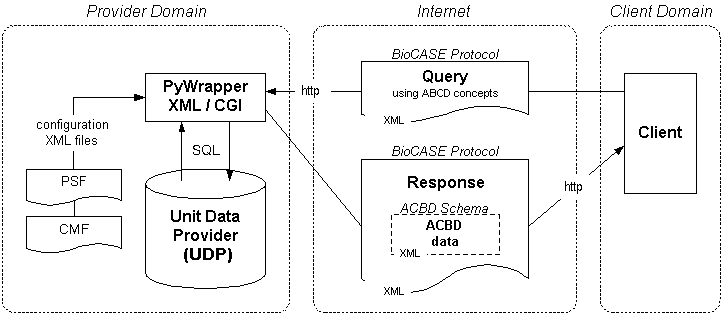
Primary biodiversity data repositories are very heterogeneous in respect to database management systems used and their data models. With the BioCASe Provider Software, aka PyWrapper, these different data sources are equipped with a common data interface that hides the implementation details of the respective repository: The data are published as a BioCASe-compliant web service, meaning all records served by a data provider can be retrieved through the usage of simple BioCASe protocol requests. This allows a network to access all data providers in the same manner, regardless of whether it's kept in a hand-tailored Access database or an off-the-shelf client/server collection management system.
Data standards and protocols
In BioCASe-based networks, biodiversity information are stored and sent across networks as ABCD documents (Access to Biological Collections Data, an XML data schema) or DarwinCore files. These documents can be retrieved through the web service by repeated BioCASe protocol requests, or, alternatively, by downloading an archive that stores all records published by a given data source. It is up to the curator of the collection or database to decide which information should be published: In particular, it's possible to exclude certain records or data items from publication (e.g. in case there are doubts about data quality or the data concern endangered species).
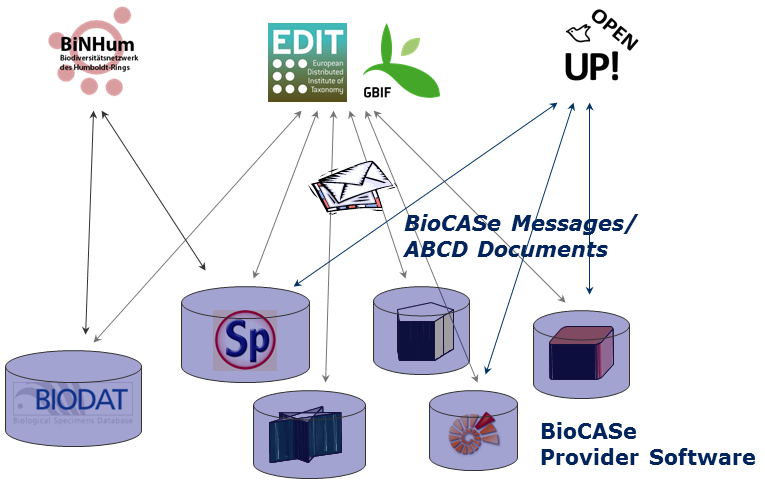
One interface serves multiple networks
Data Networks using BioCASe
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
VH/de |  |
Once fitted with this interface, a data source can be connected to a bunch of biodiversity networks. The box to the right shows the current networks for different biological communities. The smallest networks cover half a dozen providers, whereas the larger ones draw their data from hundreds of different data providers.
Getting connected can be easily done if a web server is at hand. Detailed requirements, a beginner's guide, step-by-step tutorials for installation and configuration as well as a comprehensive FAQ list are available at the PyWrapper Wiki. If you want to share you data with any of these networks, please get in contact with the Helpdesk, we're happy to assist you in the process.
