Role of the protocol
Built into the BioCASe Provider Software is the BioCASe Protocol, which defines the rules for querying a BioCASe web service. This hard-coded protocol is responsible for a successful communication between the Provider Software and any client application (usually a network component). The protocol is schema-agnostic, meaning it it indepent of the payload data that is queried and transmitted. It defines certain activities or methods that are understood by the wrapper and the client applications.
The following diagram shows the central role of the protocol and how the main components interact.
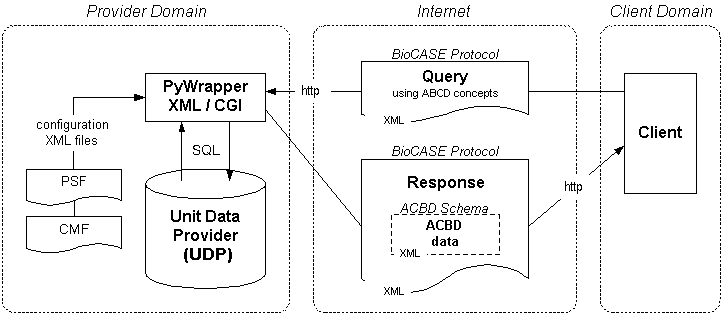
As mentioned above, the BioCASe protocol is schema-agnostic, so it can be used in conjunction with any custom-defined XML schema. In contect of the current biodiversity networks, this is usually the ABCD schema (Access to Biological Collections Data) or one of its extension schemas like ABCD-EFG (Extension For Geosciences) or ABCD-GGBN (Global Genome Biodiversity Network). Read more on schemas...
The request methods
The protocol defines several different activities in the <type> element of its header section:
<?xml version="1.0" encoding="UTF-8" ?>
<request xmlns="http://www.biocase.org/schemas/protocol/1.3" ...>
<header>
<type>search</type>
</header>
...
</request>
The BioCASe protocol defines three types of requests: Search, Scan and Capabilities:
Capabilities
A capabilities request allows a client to get information about the schemas supported by the web service and the mapped concepts. This request type returns a list of xpaths identifying all mapped concepts.
<request>
<header>
...
<type>capabilities</type>
</header>
</request>
Take a look at the examples to see a capabilities response document.
Scan
A Scan request takes one concept referenced by an xpath as an argument and returns a list of all distinct values for this element found in the database (technically, it's the result of a SELECT DISTINCT query). Here is the abbreviated scan request that would return a list of scientific names found in the database:
<header>
...
<type>scan</type>
</header>
<scan>
<requestFormat>http://www.tdwg.org/schemas/abcd/2.06</requestFormat>
<concept>/DataSets/DataSet/Units/Unit/Identifications/Identification/Result/
TaxonIdentified/ScientificName/FullScientificNameString</concept>
</scan>
Search
This is the main method. It allows to retrieve all data published for a given data schema. It can be used to get all records (by paging through the results) or just the records that match a given filter. The example below, for example, will return the first 500 records with the taxon's scientific name starting with "Abies":
<request>
<header>
...
<type>search</type>
</header>
<search>
<requestFormat>http://www.tdwg.org/schemas/abcd/2.06</requestFormat>
<responseFormat start="0" limit="500">http://www.tdwg.org/schemas/abcd/2.06</responseFormat>
<filter>
<like path="/DataSets/DataSet/Units/Unit/Identifications/Identification/Result/
TaxonIdentified/ScientificName/FullScientificNameString">Abies*</like>
</filter>
<count>false</count>
</search>
</request>
The concept schema the search filter is based on is defined by the element <requestFormat>. The concept schema to be used to return the result is defined by <responseFormat> and does not have to be the same as the requested format. For example you can query using the Darwin core standard but return a full ABCD document.
The search is stateful. So it is possible to limit the number of returned records and to define a record number to start from.
The <filter> wraps the WHERE clause of an SQL statement. It is a nested structure using different combinations of logical and comparison operators. Following operators are supported (XML is case sensitive!):
binary comparison operators
- equals
- notEquals
- lessThan
- lessThanOrEquals
- greaterThan
- greaterThanOrEquals
- like
unary comparison operators
- isNull
- isNotNull
comparison operators for multiple arguments
- in
unary logical operators
- not
binary logical operators
- and
- or
Here is a more complex filter example illustrating several conditions based on ABCD:
<filter>
<and>
<like path="/DataSets/DataSet/Units/Unit/Identifications/Identification/Result/
TaxonIdentified/ScientificName/FullScientificNameString">Abies*</like>
<or>
<like path="/DataSets/DataSet/Units/Unit/Identifications/Identification/Result/
TaxonIdentified/HigherTaxa/HigherTaxon">Pinace*</like>
<and>
<like path="/DataSets/DataSet/Units/Unit/Gathering/Country/Name">*Russia*</like>
<greaterThan path="/DataSets/DataSet/Units/Unit/Gathering/DateTime/ISODateTimeBegin">
2002-04</greaterThan>
</and>
</or>
</and>
</filter>
The response will deliver an XML document based on the protocol which envelopes an XML document based on the concept schema containing the records (see diagram above). It also gives information about the number of returned records, the number of dropped records (that is records that matched your request filter, but did contain sufficient data to produce a valid XML document based on the concept schema) and the number of records in the database that matched the query.
Take a look at the examples to see a scan response document.
Full XML Examples
search request example using ABCD as the concept schema.
search response example using ABCD as the concept schema.
scan request example using ABCD as the concept schema.
scan response example using ABCD as the concept schema.
capabilities request example
capabilities response example
