Description
The BioCASe UI is a search portal that can be used to query European
ABCD and Darwincore providers. It has been developed within
the Synthesys European project,
as a new portal to access European data. The portal is based on the
prior work done by the BioCASe interface project. You can also try
this portal (retired).
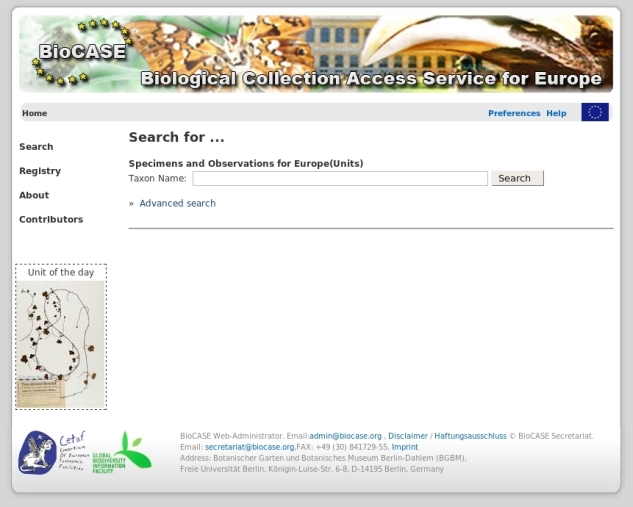
The BioCASe/EDIT portal provides two kinds of searches:
- A "simple search" allows you to perform a search based on scientific names.
- An "advanced search" allows you to perform a search based on scientific names, common names, collector names, or family names. It also enables you to restrict your search by choosing institutions, collections, countries, localities and multimedia contents.
Requirements
- Some basic system/networking administration knowledge
- Minor Python knowledges would be a plus
- A web server (Apache, IIS ...) with administration privileges and the mod_python module if you use Apache (with libapache2 for Linux)
- A units database, structured like the SYNTHESYS slide databases (contact support [at] biocase.org to get a sample or find out more.)
- Some tables for the suggest feature (optional)
- The following packages have to be installed on the server:
- Python (version 2.4 or higher)
- Cherrypy, a pythonic web application framework (2.2 <= version <= 3.0)
- Kid templates (version >= 0.9.2)
- Elementtree and setuptools; these should installed automatically with Kid
- psycopg, a python module used to query Postgresql databases
- pymssql, a python module used to query MS-SQL databases
- MySQLdb, a python module used to query MySQL databases
- pyexcelerator, a python module used to generate Excel files
- libxml and libxslt
- Feedparser to handle RSS feeds
Installation
- First, modify the file site.py (in your python path, ie /usr/lib/python2.4/site.py or C:\Python24\site.py): replace the line 364 with encoding = "utf-8" (instead of encoding = "ascii")
- Get the portal sources from our svn repository. Rename the folder (choose your own name for the portal) and move it to your favourite directory on the web server.
- Open a command prompt and go to the directory where you just placed the sources. On Linux/Mac run "python setup.py", on Windows "<PythonPath>python.exe setup.py".
- Answer each question of the setup script to set the different parameters; you will be able to change/correct them later in the configuration files if needed. The script will check for the required packages and direct you to the pages you can get them in case they're not installed.
(Hints: Choose "no" for the question "Please indicate if the portal is for taxonomist" if you don't want to use a thesaurus database; Choose "no" for "Do you have a database with metadata, similar to the CORM database" if you don't have one.) - After the script is completed, update your web server's configuration file with the configuration lines given by the script and restart your server.
- Test your installation (URL will be displayed at the end of the configuration script).
Configuration
The configuration parameters can be changed in the following files (path/filename):- webapp/configfile.cfg - Cherrypy and general application settings; you should not change anything except the URLs of the XSLTs.
- config/admin.ini - specifies if the portal is for taxonomists or not and what sort of DBMS you're using (MS SQL Server or MySQL).
- config/config.ini - Mail server properties, the portal's name and location, location for thesaurus configuration files.
- config/logconf.ini - Logging properties.
- references.xml - List of thesauri with their official names, displayed names and displayed references. The boolean "reference display='True'" specifies if the thesaurus should be used for this portal installation.
- relations.xml - List of relations known to the different thesauri.
- thesaurusList.xml - List of available thesauri for the taxonomist's portal version; type=name of the thesaurus (must match the configuration file name in TOQE) and the TOQE URL to access the service.
- thesaurusListSimple.xml - List of available thesauri for the simple version (author names and concepts will not be displayed); type=name of the thesaurus (must match the configuration's file name in TOQE) and the TOQE URL to access the service.
#######
#portal
#The values of PythonInterpreter don't really matter, as long as they're different between the two Location blocks.
######
<Location /appName>
SetHandler mod_python
PythonPath "['yourPath/appName']+['yourPath/appName/webapp']+['yourPath/appName/lib']+['yourPath/appName/log']+sys.path" PythonInterpreter appName
PythonHandler mpcp
PythonOption wsgi.cleanup cherrypy::engine.stop
PythonOption cherrysetup serverStarter::configureServer
PythonOption sys.path 'yourPath/appName/lib'
PythonAutoReload Off
PythonDebug Off
</Location>
For the suggestion tool (Note: PHP must be installed on the server denoted by 'anURL'; an example of suggest.php can be found in /appName/webapp/static/php/dojotree.):
ProxyRequests off
ProxyPass /suggest/ http://anURL/php/suggest/suggest.php
ProxyPassReverse /suggest/ http://anURL/php/suggest/suggest.php
Known Problems
If you are using Python2.6, you might have to edit the __init__.py file from Kid:def import_template(name, encoding=None):
"""Import template by name.
This is identical to calling `enable_import` followed by an import
statement. For example, importing a template named foo using the normal
import mechanism looks like this:
import kid
kid.enable_import()
import foo
This function can be used to achieve the same result as follows::
import kid
foo = kid.import_template('foo')
This is sometimes useful when the name of the template is available only
as a string.
"""
enable_import()
# mod = __import__(name)
## hack for python2.6
filename = name
name=os.path.splitext(filename)[0]
parts=name.split(os.path.sep )
mod,name=".".join(parts),parts[-1:]
mod = __import__(mod,globals(),locals(),name)
components = filename.split('.')
###
# components = name.split('.')
for comp in components[1:]:
mod = getattr(mod, comp)
if encoding:
mod.encoding = encoding
return mod
It has been tested on Windows XP Professional (SP 2), Debian 3.1 ,
and Ubuntu 6.06 LTS-8.10.
As client browsers, Internet Explorer, Mozilla Firefox, Opera and Safari have been found OK.
